Applying Independent Component Analysis (ICA) on Poly5 data.
EEG signals can contain unwanted physiological artifacts, such as eye movements. Researchers often find wish to remove these artifacts in post-processing of the data. This Knowledge Base article describes how eye movements can be removed from EEG signals acquired using TMSi amplifiers with independent component analysis (ICA). Read this related blog for general information on the ICA technique and how it works.
TMSi offers a Python interface for APEX and SAGA amplifiers, allowing users to record EEG data and save it in a Poly5 file format. This interface can also be used for post-processing as it enables users to read the Poly5 file. Moreover, it provides an example of how to remove eye movements from the EEG data with ICA. Applying ICA for artifact removal consists of three phases. First, the data is converted from Poly5 to an MNE object with the correct properties, next the data is observed and made ready for the ICA. The final step is the ICA process itself.
The example script that is described in this Knowledge Base article is named 'example_ica.py'. To run this script, the example data set 'sample_eeg_eog_ecg_emg.Poly5' can be used. This data set contains EEG channels along with additional bipolar EOG, ECG and EMG channels. This example script and data can be found here.
Step 1: Poly5 to MNE
Loading the Poly5 data to an MNE object can be done using the Poly5Reader in the above mentioned example script. As the Poly5 data format does not contain meta-data regarding channel locations and channel type, this information has to be added manually. Adding the channel locations for the EEG channels is necessary to be able to analyze the different ICA components later. Channel types of unipolar channels are automatically set to ‘eeg’ in the Poly5Reader. In case bipolar or auxiliary channels are used, for example, to measure eye movements seperately (EOG signals), their channel types have to be set manually in order for MNE to use them correctly.
Step 2: Observe and clean data
Once the data is loaded and converted, the data can be analyzed in MNE. The first step is to plot and observe the data. This is done to determine the required pre-processing. Before applying ICA, the removal of bad channels and bad data spans should be handled first. Additionally, the data should be high-pass filtered to get rid of the baseline drift which might seriously influence the quality of the ICA decompositions. In this step, one should also judge whether ICA is required and the right tool for artifact repair.
Step 3: Apply ICA
The next step is to use ICA to decompose the data into its separate independent components. The different components can then be analyzed either manually or automatically and the components representing the artifacts can be selected for exclusion. More information about visually identifying components can be found in the EEGlab turtorial. Finally, applying the ICA results, with the artifact components excluded, generates the signals of the different electrodes on the unfiltered data with the artifacts removed.
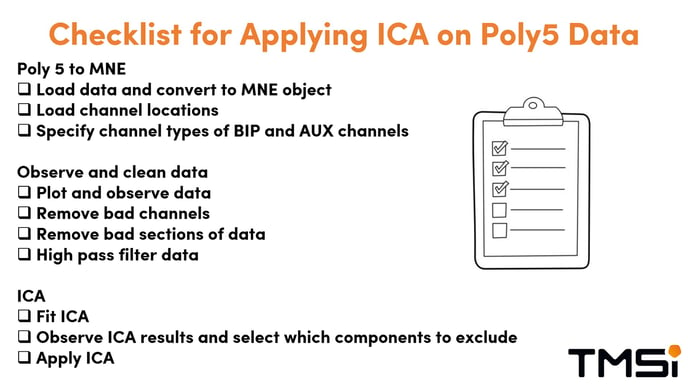
In the figure below, all steps needed to apply ICA on Poly5 data is summarized.
Do you have further questions on this topic?
Contact ✉️ support@tmsi.com, 📞+31 (0)541 539 013 or 📄 request support.